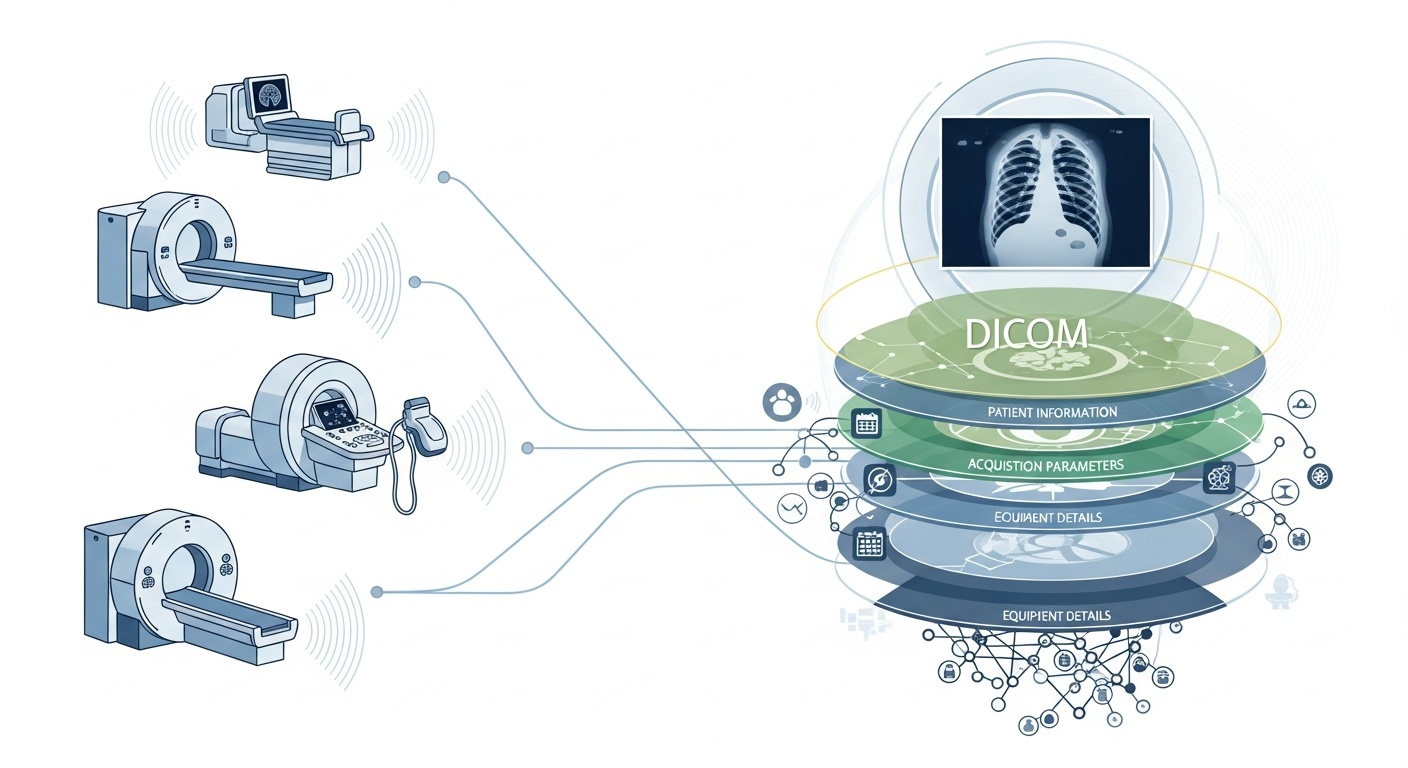
Every time a radiology team scans a patient, say a CT, MRI, or ultrasound, a cascade of data is generated. Most visible is the image itself, but behind it is a rich layer of metadata: the who, what, when, how, and where of the scan. That layer is governed by the standard DICOM (Digital Imaging and Communications in Medicine), a medical-imaging format standard baked in by the National Electrical Manufacturers Association (NEMA) and the American College of Radiology (ACR) decades ago.
What makes the metadata so interesting is that it is structured, machine-readable, and remarkably detailed: equipment settings, acquisition parameters, patient demographics, study IDs, even institution codes and modality manufacturer details. That richness is what enables big-data analytics, downstream research, AI modelling, and protocol standardisation if you harness it well.
In the context of big data, we’re not just talking about a few dozen imaging studies. We’re talking hundreds of thousands, or even millions, of images across modalities, sites, and vendors, with metadata as the key indexing layer: the “who & when & how” of each item. Without effectively utilising metadata, you risk having a massive archive of images but minimal ability to query, compare, or derive insight from them. A recent review states: “the majority of information stored in PACS archives is never accessed again,” a wasted opportunity.
Say you’re running a multi-site study of lung CT scans for early emphysema detection. You’ll want to select scans based on parameters: scanner vendor/make/model, slice thickness, reconstruction kernel, patient age, date ranges, perhaps even dose parameters. Most of those are metadata fields, not pixel data. Extracting those tags enables you to build the cohort, exclude incompatible scans (e.g., too-thick slices), and ensure comparability.
Metadata lets you monitor the imaging process itself: is the institution using the correct protocol? Are acquisition parameters deviating over time (e.g., field of view, contrast injection timing)? Are the vendor settings consistent? In a big-data world where thousands of scans happen per day, you cannot rely on human eyeballing. You need analytics on metadata. Many PACS systems under-exploit metadata for this reason.
If you build an AI or radiomics pipeline, you cannot treat every image as interchangeable. Metadata becomes an integral control variable: input features often include modality, kVp (kilovoltage peak), manufacturer, kernel, slice thickness, and even the date or hospital may matter (domain shift). These metadata fields help manage bias, harmonise data, and annotate images with context. Many researchers call metadata “as important as pixel data.”
Big data means scale. That implies varied sources, multiple vendors, different institutions, and heterogeneous formats. DICOM metadata is the standardised “language” that helps unify the metadata layer, enable search/index, ensure interoperability, and build scalable architectures (cloud-PACS, federated archives). But implementation matters. The same PMC study found that many systems don’t fully exploit the standard.
PostDICOM offers a cloud-based PACS (Picture Archiving and Communication System) for storing, viewing, and sharing imaging studies and clinical documents. Some key metadata-relevant features of PostDICOM:
• Support For Dicom Tags & Descriptions: Our resource library lists “DICOM Modality & Tags,” allowing users to access tag lists and descriptions.DICOM Modality & Tags
• Api / Fhir Integration: It supports API and FHIR (Fast Healthcare Interoperability Resources) interfaces, allowing metadata to be programmatically queried, integrated with other systems, and analysed.
• Cloud Scalability & Multi-site Sharing: Sharing across patients, doctors, institutions; unlimited scalability means big-data pipelines become feasible.
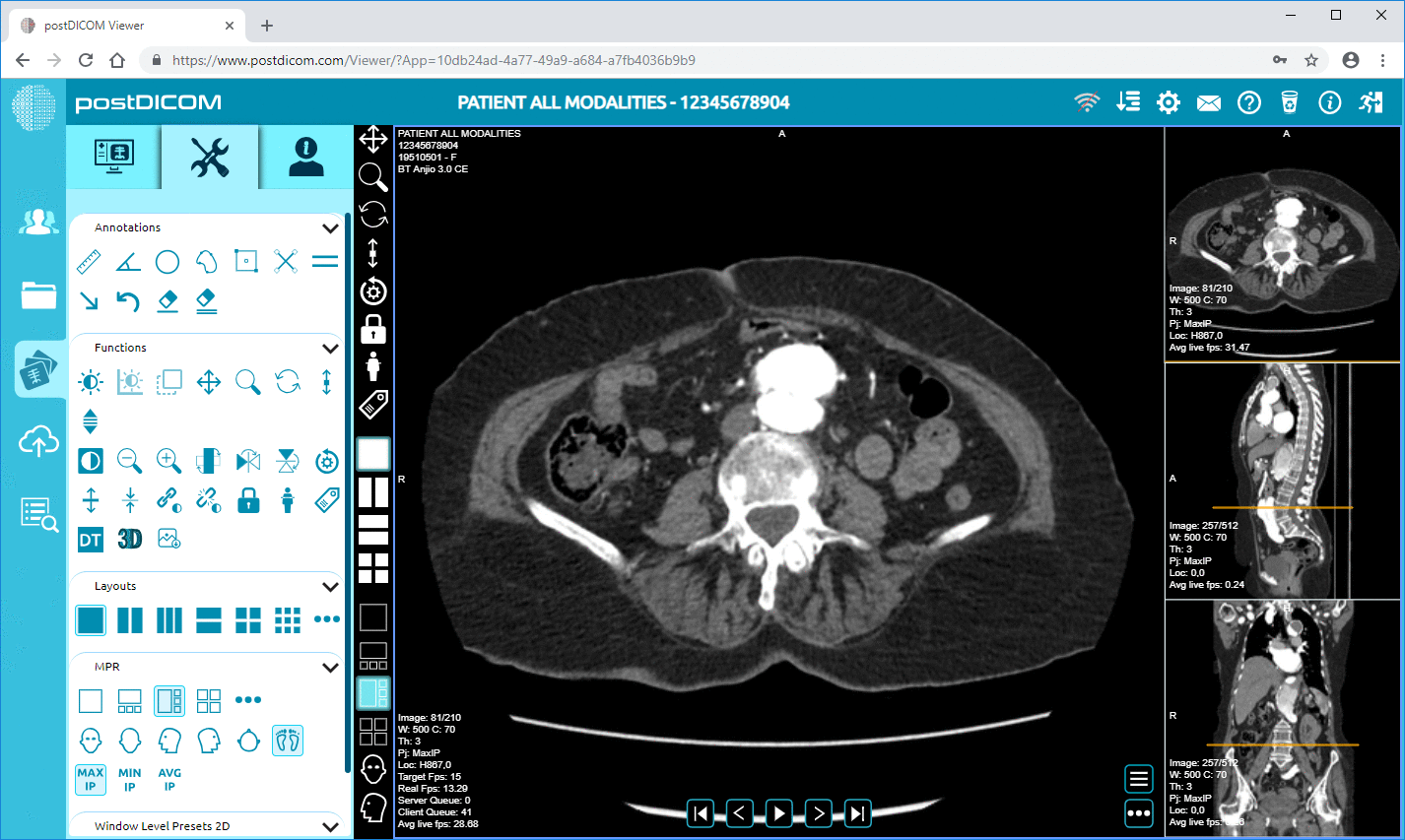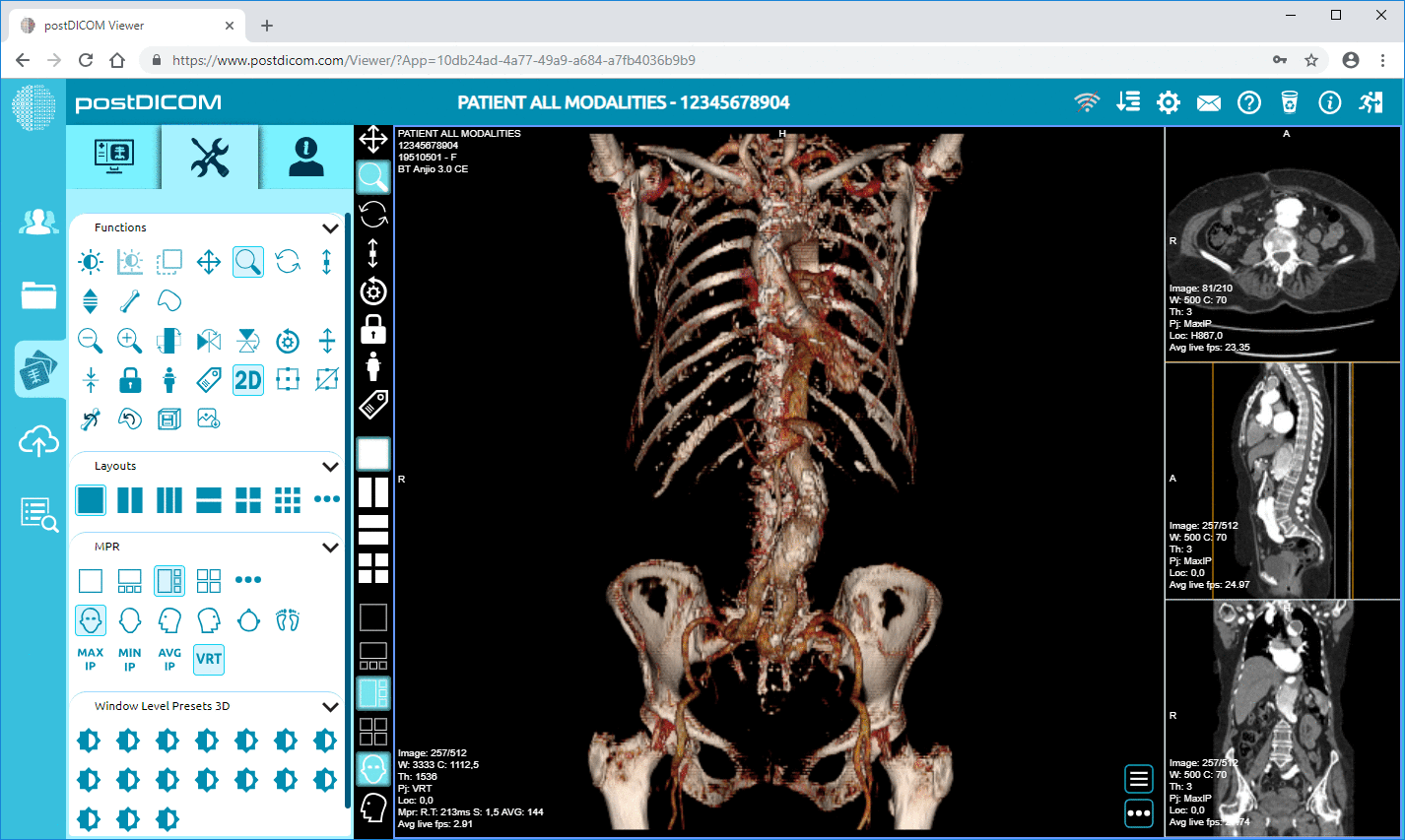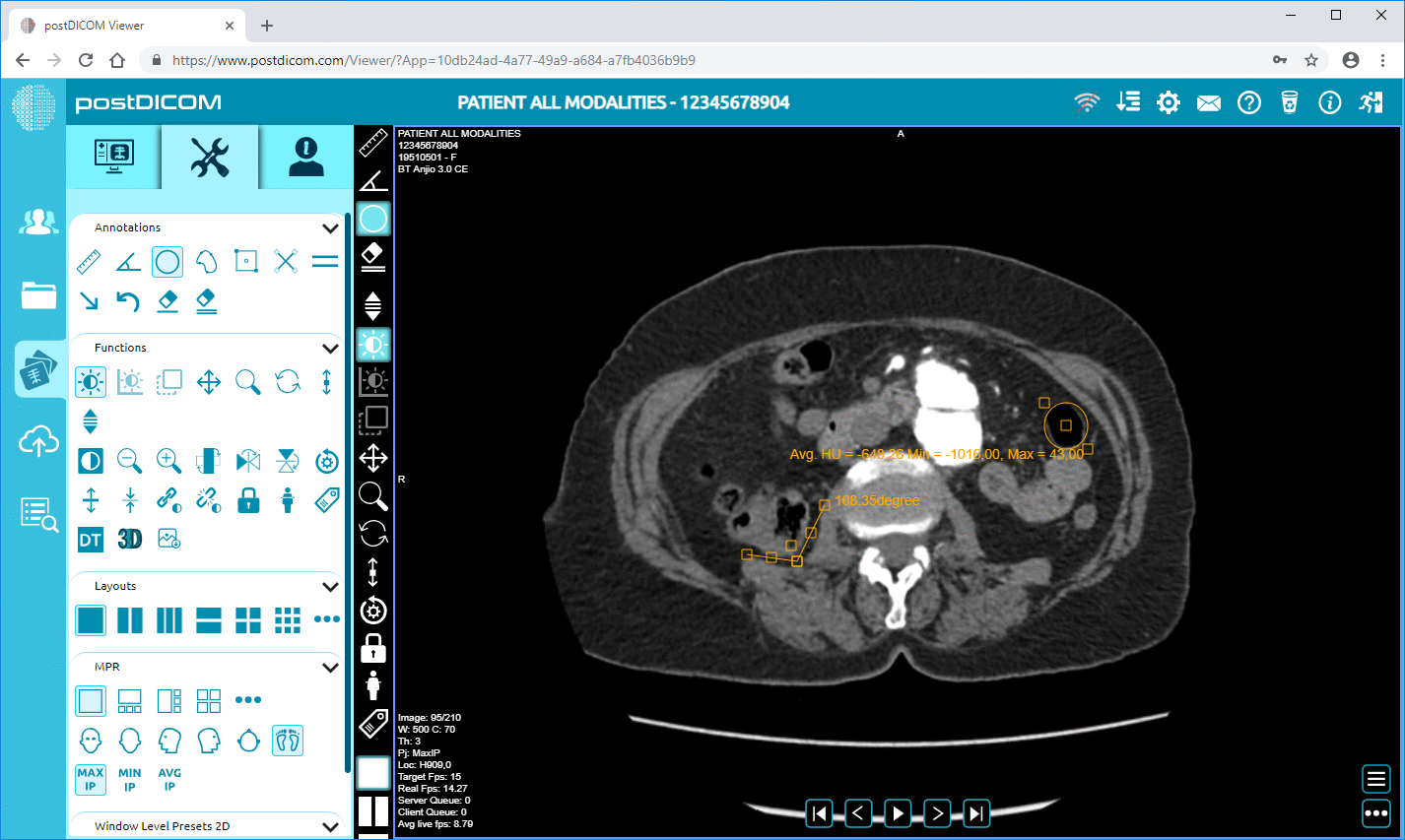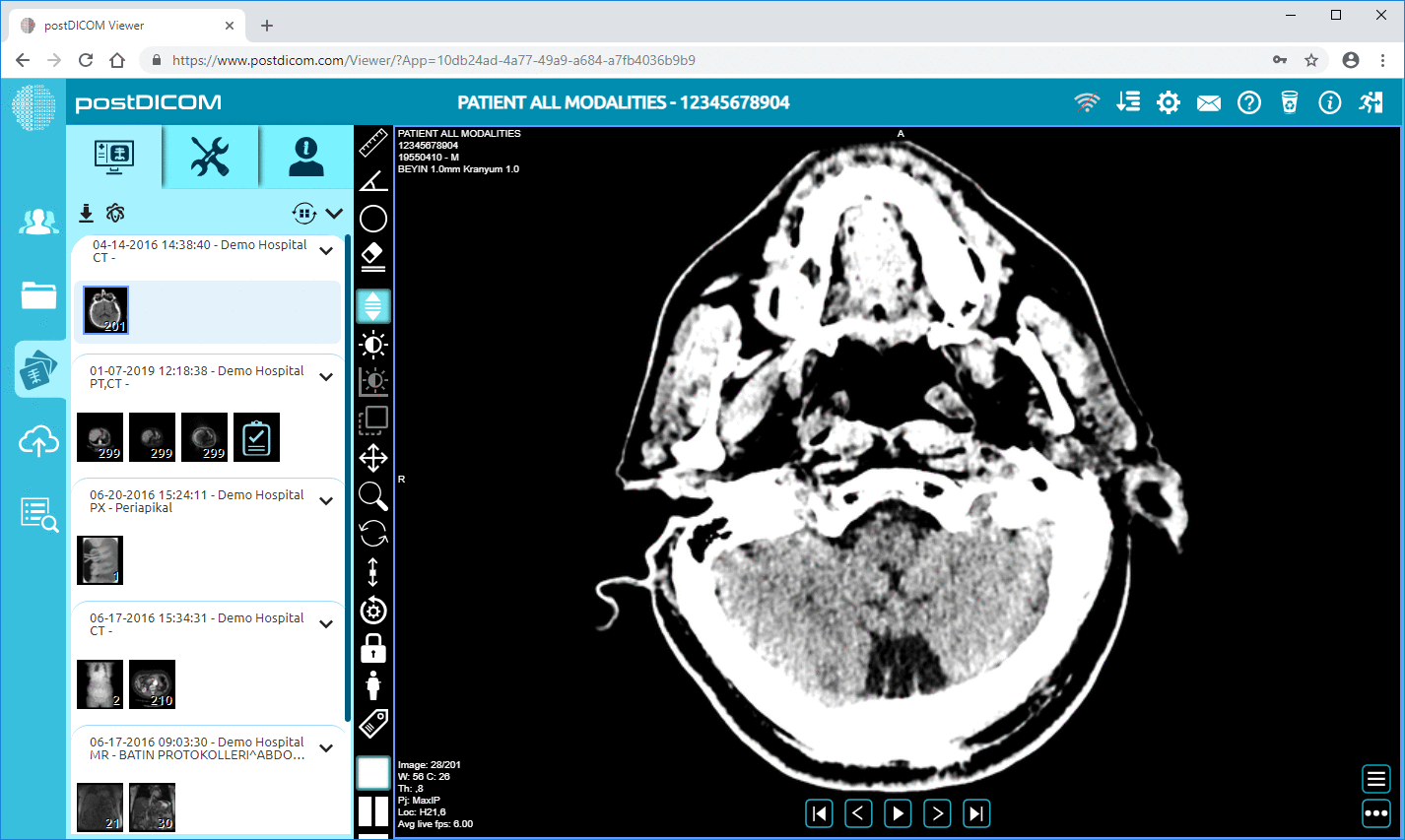
• Advanced Image Processing & Multi-modality Support: While this concerns pixels, the support of modalities like PET–CT and multi-series means the metadata is substantial (SUV values, fusion volumes, modality type) and available for analytics.
Using a platform like PostDICOM allows you to harness metadata through structured workflows, APIs, and cloud architecture.
Here’s how to structure the workflow from raw archives to analytics-ready insights.
 - Created by PostDICOM.jpg)
The first step in harnessing DICOM metadata for analytics is extraction and normalisation. Libraries such as the open-source Python package PyDicom are commonly used to parse DICOM files and extract relevant tags, including image rows and columns, convolution kernels, and modality-specific acquisition parameters.
Handling heterogeneity is crucial, as different vendors often use private tags or non-standard implementations. Robust parsing, fallback logic, and comprehensive tag-mapping tables are required to ensure consistency across datasets.
Once extracted, the metadata must be normalised and mapped to standard ontologies and structures, such as modality codes, vendor names, slice thickness categories, and standardised date and time formats.
Finally, the structured metadata should be stored in a big-data environment, such as a relational database, NoSQL store, or columnar data lake, with indexing to enable fast, efficient querying.
Once extracted, metadata must undergo quality assurance to ensure accuracy and reliability. Missing or inconsistent fields, such as blank slice thickness values, inconsistent modality labels, or duplicate Study Instance UIDs, need to be identified and corrected.
Privacy and anonymisation are also critical at this stage, as metadata often contains personally identifiable information including patient names, IDs, and dates; de-identification tools and protocols are essential.
Maintaining comprehensive audit trails is another important practice, documenting when metadata was extracted, which parser versions were used, and any errors encountered during the process.
Governance policies should also define mandatory fields and provide guidance on how to handle legacy or incomplete datasets to ensure that downstream analytics are accurate and compliant.
The next step is metadata-driven indexing and feature engineering, which transforms raw metadata into actionable information.
This involves creating indices and filters that allow researchers and analysts to query specific datasets, for example, retrieving all chest CT scans with slice thickness below 1.5 millimeters from a particular vendor within a given date range.
Feature engineering builds upon this by combining metadata fields such as vendor, model, acquisition date, slice thickness, convolution kernel, contrast protocol, body region, radiation dose, and institution ID into structured variables suitable for analysis.
Metadata can also be linked to clinical datasets, connecting imaging data to patient outcomes, diagnoses, or treatments. This linkage allows a more holistic view of imaging data and its clinical context.
Once metadata is indexed and features are engineered, analytics and insight generation become possible.
Descriptive analytics can reveal study volumes by modality, vendor, or region, track trends in acquisition parameters, and highlight errors or inconsistencies in imaging practices. Comparative analytics enable evaluation of acquisition protocols across institutions, detection of deviations, and identification of outlier scans that may require special attention.
For machine learning and AI applications, metadata is essential for controlling domain shift, ensuring that training and test datasets are stratified appropriately, and combining pixel-based features with structured metadata variables. Operational dashboards can then leverage this data to monitor workload, assess quality assurance metrics, and ensure protocol compliance across sites.
Finally, feedback and continuous improvement complete the metadata lifecycle. Insights derived from analytics can inform the refinement of acquisition protocols and standardisation of workflows to improve overall data quality.
New imaging studies and metadata should be continuously ingested, with monitoring of the metadata store’s performance, query times, and data integrity. Lessons learned should be archived to capture predictive metadata fields, address recurring gaps or errors, and improve governance practices.
This iterative approach ensures that metadata pipelines remain robust, scalable, and valuable for future research, AI applications, and operational decision-making.
• Vendor/institution Variability: Private tags or loose standard interpretations.
• Missing Or Corrupted Metadata: Older studies may have incomplete headers.
• Data Privacy & Anonymisation: PHI must be de-identified for multi-site research.
• Scale & Performance: Millions of images require efficient processing and storage.
• Domain Shift/bias: Dominant vendors/protocols may skew AI models.
• Regulatory & Compliance Issues: Multi-region deployments may involve HIPAA, GDPR, or local regulations.
DICOM metadata is the hidden skeleton of imaging analytics. Platforms like PostDICOM illustrate how to transform a fragmented archive of DICOM files into a searchable, scalable, metadata-driven ecosystem. If you want to explore PostDICOM, we encourage you to get our 7-day free trial.
|
Cloud PACS and Online DICOM ViewerUpload DICOM images and clinical documents to PostDICOM servers. Store, view, collaborate, and share your medical imaging files. |